The Restriction endonucleases recognizes a specific base sequence of four to eight bases in double-stranded DNA and cleaves both strands of the duplex.
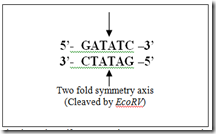
There are known type of restriction endonucleases : type I,II and III. Type II enzymes are frequently used in the rDNA technology. Type I and type III are not use because these type enzymes cleaves the DNA far from the recognition sites. The Restriction endonuclease recognizes particular specific sequences are 4 to 6 nucleotides recognize by the type II enzymes because these enzymes only cleave at this site. The palindrome sequence is “Two-fold symmetry” as shown below.
The type II restriction rnzymes are discovered and characterized by “Hamilton Smith” and “Daniel Nathans” in 1960. Approximatly 200 Restriction endonucleases are isolated from bacterial species like- E.coli, Bacillus, Haemophillus, Streptococcus and thermus aquaticus.
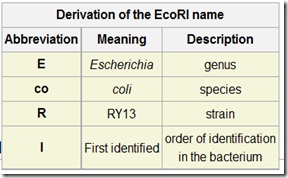
Restriction endonucleases is named by the first letter of the “genus” of the bacterium that produced it and the first two letters of its “species”, followed by its serotype (or) strain designation, if any, and a roman numerical if the bacterium contains more than one type of restriction enzymes.
E.g.: EcoRI is produced by “E.coli” strain RY13
Bam HI isolated from “Bacillus amyloliquefaciens”
Bgl II isolated from “Bacillus globigis”
Hae III isolated from “Haemophilus aegyptius”
Pst I isolated from “Providencia stuartil”
No comments:
Post a Comment