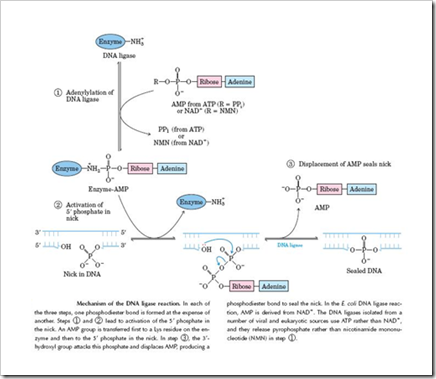
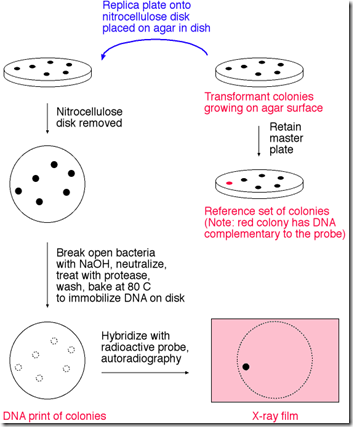
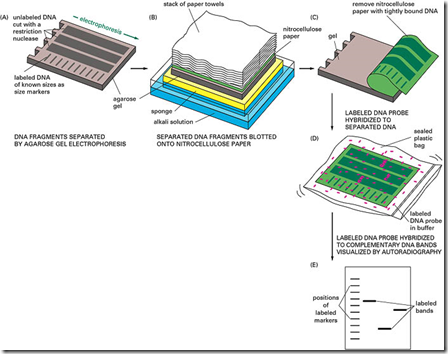
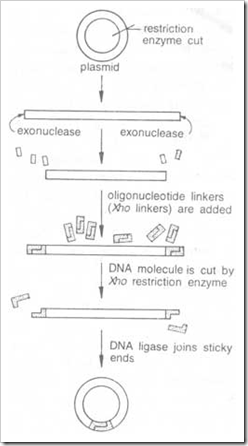
Introduction:
The DNA fragment (or) the gene of interest can be linked to a carrier molecule, which can transport the gene of interest into the host cell. This carrier molecule is referred to as a “Cloning vector” (or) “Cloning vehicle”; the cloning vehicle is the central component of a gene cloning experiment and it constitutes the gene transfer system.
Important features of vectors:
-
It must be able to replicate.
-
There must be some way to introduce vector DNA into a cell.
-
There must be some means of detecting its presence, preferably by a plating test.
-
It should contain an assortment of unique Restriction endonucleases cleavage sites.
-
It should occur in large number of copies.
Type of vector:
There are three main types of vectors in use. They are
1) Plasmids
2) Cosmids
3) Phagemids
1) Plasmids:
Plasmids are extra chromosomal, autonomously replicating , small circular molecules of DNA found in many prokaryotes and in a few eukaryotes such as the yeast “Saccharomyces cerevisiae”.
Properties of Plasmids:
-
They replicate independently (or) autonomously
-
Most of them are circular duplex of DNA molecules.
-
They have an origin of replication naturally in them
-
They are passed on to the daughter cells during cell division.
-
They may carry very important genes for antibiotic resistance, toxin production, for antibody production, for degradation of a large number of unusual substrates such as herbicides (or) industrial effluents and genes for nitrogen fixation. These confer the phenotypic traits of plasmids.
-
They rely on the DNA replication enzymes of the host cells for their replication; however, the initiation of replication is controlled by plasmid genes.
-
Certain plasmids do not show any phenotypic traits such as plasmids are called “Cryptic plasmids”.
-
They have high transformation efficiency.
-
They have convenient selectable markers such as antibiotic resistance, toxin production etc, for transformants and recombinants.
-
They have the ability to clone reasonably large pieces of DNA say about 5 kilo base pairs.
-
They are of low molecular weight.
-
They are easily isolated and purified.
Size of plasmids:
Plasmids are duplex, supercoiled DNA molecules and they range in size from 1X106 Daltons to greater than 200X106.
Number of plasmids:
The number of copies of plasmid in a cell is referred to as “Copy number”. When there are one (or) two copies, the copy number is called “low copy number”. When there are twenty (or) more copies per cell, the copy number is called “high copy number”
Plasmid classification:
The naturally occurring plasmids are classified based on the main characteristics coded by the plasmid genes. It is grouped into FIVE main types:
a) F-plasmids
b) R-Plasmids
c) Col – plsamids
d) Degradative plasmids
e) Virulence plasmids
a) F-Plasmids (Fertility plasmids):
These plasmids carry only “tra” genes (transfer gene) and no characteristic beyond the ability to promote conjugate transfer of plasmids. The presence of “tra genes” promotes bacterial conjugation. These plasmids may be denoted as F+ and F-, which means those having the fertility (F) factor and those without it. These plasmids are not used in gene cloning. Most of the “tra genes” are involved in “pili synthesis” (sex pili) on donor.
b) R-Plasmids (Drug resistance):
These carry genes conferring on the possessor resistance to one (or) more antibacterial agents such as “Chloramphenicals”, “Amphicillin”, “Tetracycline” and any metal. The “R” strands for “Resistance”. “Plasmid RP4” found in pseudomonas is an example of R-plasmid. This R-factor was discovered in Japan in 1955.
The R-factor is wide spread in contain strain of almost all pathogenic bacteria. The plasmid genes after encode for enzyme that chemically inactivate the drug (or) by active export eliminate it from the cell.
c) Colicinogenic (or) Col plasmid:
Col plasmids are E.coli plasmid able to produce colicins, proteins that prevent growth of susceptible bacterial strains that do not contain a col plasmid. The bacterial toxins are generally called “Bacteriocin” these bacteriocin are active only against closely related strains of bacteria toxins of this. Types that are liberated by strains of E.coli are called “Colicins”. The colicins are simple proteins. Several different types of colicins have been isolated which kill sensitive cells by different mechanisms. The plasmids containing genes for such toxic substance, “colicin” is called “Col plasmid”. Depending upon the nature of colicins, there are different types of col plasmids. They are col B, colE1, col E2, col I and col V. the toxin from “Pseudomonos” is called “Pyrocins”.
d) Degradative plasmids:
These are plasmids, which have genes for enzymes that enable the bacterium to metabolize unusual substrates such as Toluene, xylene and salicylic acid. These plasmids are also called “Dissimilation plasmids”. This plasmid type (Ptol) is responsible for the ability of certain Pseudomonas species to break down different to degrade industrial solvents such as toluene and xylene. A combination of several plasmids, when transferred to pseudomonas bacteria, allows the bacteria to break down complex hydrocarbons and other compounds present in crude oil. The bacteria, containing these plasmids have a potential use for treatment of environments contaminated with oil spills.
e) Virulence plasmids:
The plasmids have genes that confer pathogenicity on the host bacterium. For example, Ti-plasmids found in “Agro bacterium tumefacience”. They include crown gall disease on dicotyledonous plant.
2) Cosmids
3) Phagemids